About StatAlign
StatAlign provides a unified approach for inferring multiple
sequence alignments, evolutionary trees and model parameters within a joint Bayesian framework.
Most other methods for phylogenetic inference use a single fixed alignment as input. However, this
can result in significant bias in the resulting trees, since the results may be very sensitive to
the specific choice of alignment. In addition, these methods typically treat
insertions and deletions (gaps) as missing data, discarding a great deal of important information
in the process, and potentially further biasing the results.
To address these problems, StatAlign jointly samples multiple alignments, evolutionary trees
and model parameters under a stochastic model of substitution, insertion and deletion
 , making use of a Markov chain Monte Carlo scheme
, making use of a Markov chain Monte Carlo scheme
 to generate samples from the desired posterior distribution.
The program allows a range of different substitution models; the insertion-deletion model
is a modification of the TKF92 model
to generate samples from the desired posterior distribution.
The program allows a range of different substitution models; the insertion-deletion model
is a modification of the TKF92 model
 , which allows 'refragmentation' at internal nodes. For more details,
see Miklós et al. (2008)
, which allows 'refragmentation' at internal nodes. For more details,
see Miklós et al. (2008)
 .
.
Users can load sequences, and choose a substitution model with which to analyze
the sequences. Once the parameters of the Markov chain and desired output are specified
(e.g. log-likelihood trace, alignment, tree samples etc.), the MCMC chain can be initialised,
generating a set of samples for the parameters of interest, as well as
consensus alignments and trees.
The following sections contain descriptions of the graphical elements of the program (menu bar,
tabulated panels, pop-up dialog windows), with examples illustrating how an analysis can be carried
out.
[
File
In the File menu, we can load sequences (and other types of data if supported by plugins),
set the output preferences and exit the program.
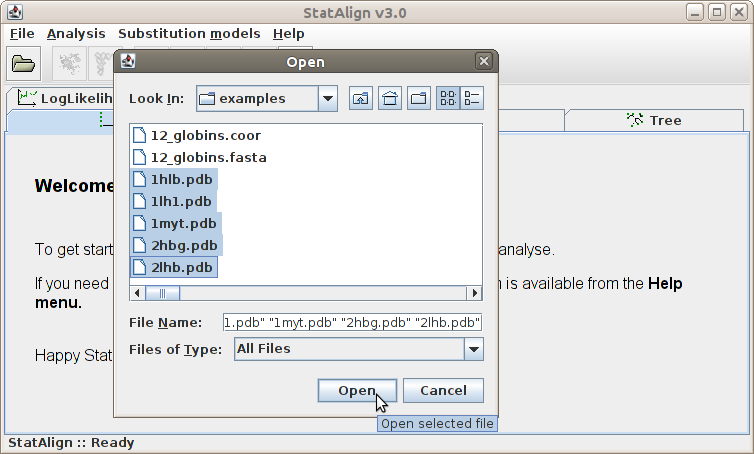
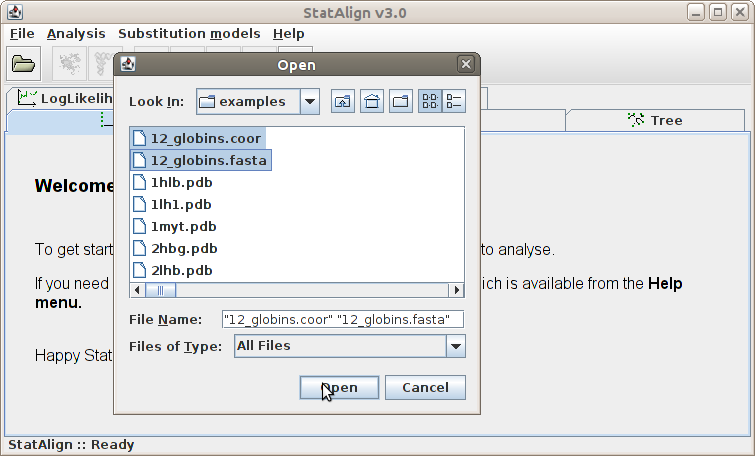
Add sequence(s)...
Clicking this menu item will result the pop up of a file opening dialog window.
We can browse the directories in this window and open a file. The file to be opened
should contain sequences in Fasta file
 format (and potentially other types of data, when appropriate plugins exist).
Sequences can be viewed and removed via the
Sequences panel.
format (and potentially other types of data, when appropriate plugins exist).
Sequences can be viewed and removed via the
Sequences panel.
Output settings...
Clicking this menu item will pop up a dialog window where
you can set up your
output preferences
Exit
Exits StatAlign.
[
Analysis
Run
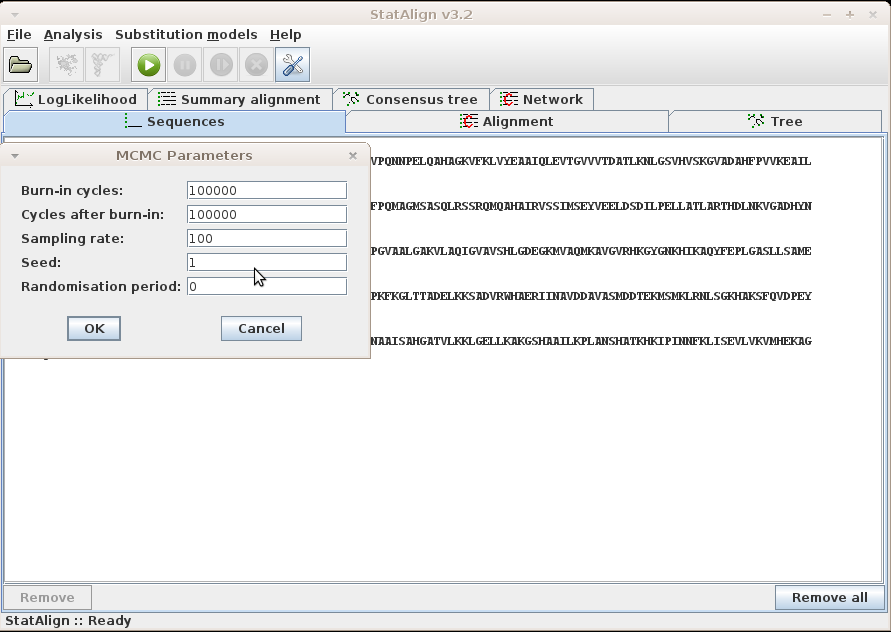
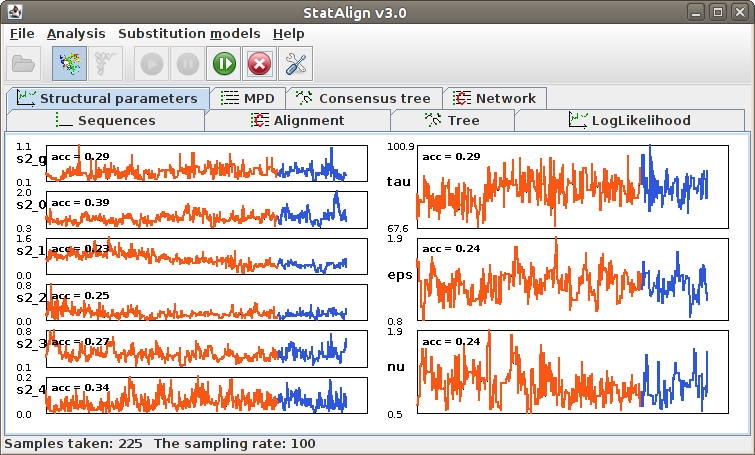
After loading your sequences and choosing your Model and Output settings, you can click this menu item to set the MCMC parameters and start the MCMC analysis.
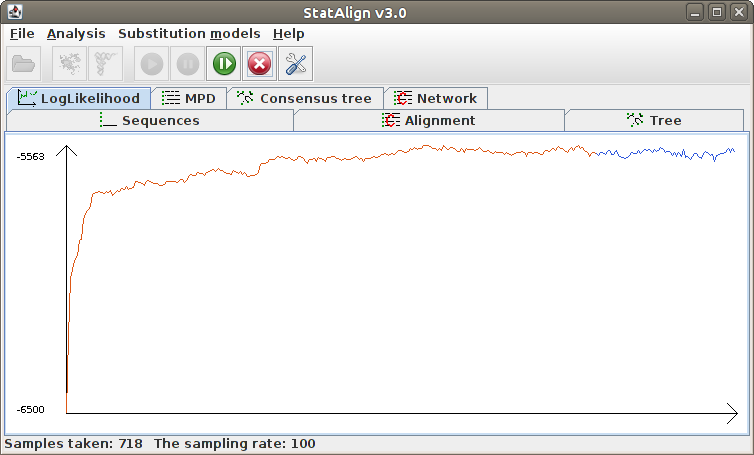
Information will be given about the progress of the MCMC sampling in the Status
bar, such as the number of burn-in steps completed and the number of MCMC samples
taken. The Postprocessing panels will show both snapshots
from the MCMC chain, as well as various summaries calculated from the MCMC samples.
Pause
We can pause an active MCMC run by clicking this button.
Resume
A suspended run can be continued by clicking here.
Stop
This item stops an MCMC run. Output files will still be created from the samples
that have been taken so far, but you won't be able to restart the process from the
same point. This should be used only if you indeed want to terminate the MCMC run early.
[
When StatAlign completes a run, it outputs files containing the results of the MCMC sampling. By default, the alignment and total log-likelihood for each MCMC sample are written to a .log file, which also contains a report of the acceptance rates for each MCMC move.
Additional MCMC output is generated by postprocessing plugins, which extract properties of interest from each MCMC sample. This output can either be added to the .log file, or sent to individual files.
| File extension | Contents |
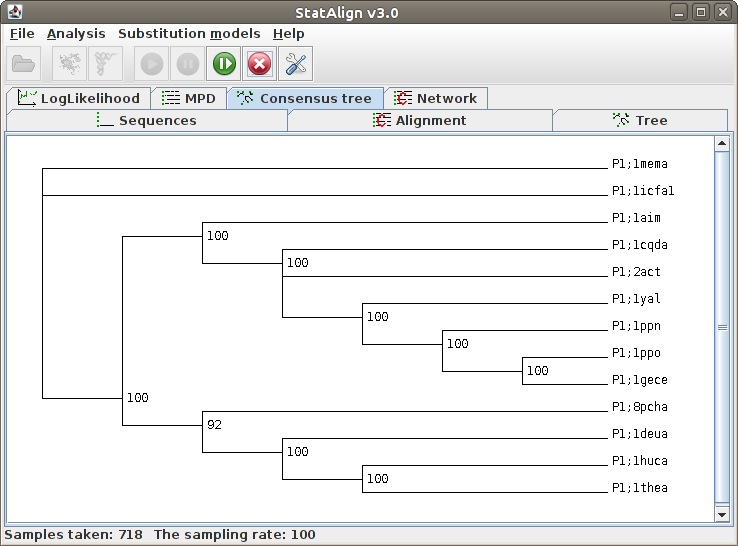
| .tree | Sampled trees (Nexus format) |
| .ctree | Consensus tree taken from samples so far. Internal nodes are labelled with posterior probability for the split preceding the node. |
| .coreModel.params | Parameters of the core evolutionary model (usually indel and substitution rates) |
| .ll | Log likelihood for each MCMC sample. The first column contains the contribution from the core evolutionary model, and the second column contains the total including contributions from model extension plugins. |
| .mpd.ali | Contains the minimum-risk (also termed maximum posterior decoding) summary alignment. |
| .mpd.scores | Per-column marginal posterior probabilities for each alignment column in the MPD alignment. |
| .ali | Sampled alignments (several possible formats). By default this file is not created, since the alignments are printed to the .log file instead. |
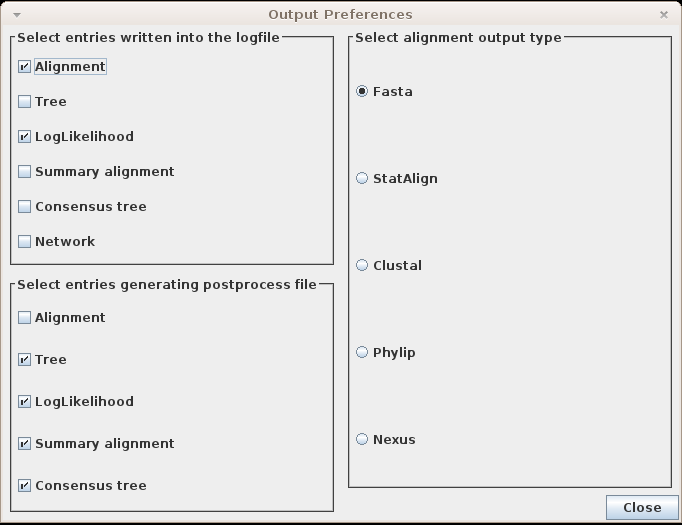
Logfile
On the top left corner of the output preferences window, users can choose which
postprocesses write into the logfile. The format of the output
is the following:
Sample [sample_number] [TAB] postprocess_name: [TAB] data
For example, selecting to output just the tree and log-likelihood to the log file, we will obtain a file containing the following:
Sample 0 Loglikelihood: -3718.11540541795
Sample 0 Tree: ((P1_1aeia:0.05535,((P1_1ann:0.10414,P1_1axn:0.04547):0.01,(P1_1ala:0.02321,P1_1avha:0.01238):0.07713):0.01):0.03021,P1_2ran:0.08613);
Sample 1 Loglikelihood: -3620.5414903951587
Sample 1 Tree: ((P1_1aeia:0.05535,((P1_1ann:0.10414,P1_1axn:0.04547):0.01,(P1_1ala:0.02321,P1_1avha:0.07235):0.07713):0.01):0.03021,P1_2ran:0.12924);
Sample 2 Loglikelihood: -3618.189925496592
Sample 2 Tree: ((P1_1aeia:0.05535,((P1_1ann:0.10414,P1_1axn:0.04547):0.01,(P1_1ala:0.02321,P1_1avha:0.11236):0.07713):0.01):0.04352,P1_2ran:0.11424);
Sample 3 Loglikelihood: -3615.7331381880526
Sample 3 Tree: ((P1_1aeia:0.05535,((P1_1ann:0.10414,P1_1axn:0.04547):0.01,(P1_1ala:0.02321,P1_1avha:0.11236):0.07713):0.01837):0.04352,P1_2ran:0.11424);
Sample 4 Loglikelihood: -3587.894867267956
Sample 4 Tree: ((P1_1aeia:0.05535,((P1_1ann:0.10414,P1_1axn:0.06347):0.01,(P1_1ala:0.02321,P1_1avha:0.11236):0.07713):0.01837):0.04352,P1_2ran:0.11424);
Sample 5 Loglikelihood: -3585.9665576563943
Sample 5 Tree: ((P1_1aeia:0.05535,((P1_1ann:0.10414,P1_1axn:0.06347):0.01,(P1_1ala:0.02321,P1_1avha:0.11236):0.08542):0.01837):0.04352,P1_2ran:0.11902);
Sample 6 Loglikelihood: -3551.4723341911595
Sample 6 Tree: ((P1_1aeia:0.05535,((P1_1ann:0.10414,P1_1axn:0.06347):0.05301,(P1_1ala:0.02321,P1_1avha:0.11236):0.08542):0.03414):0.04352,P1_2ran:0.11902);
.
.
.
[
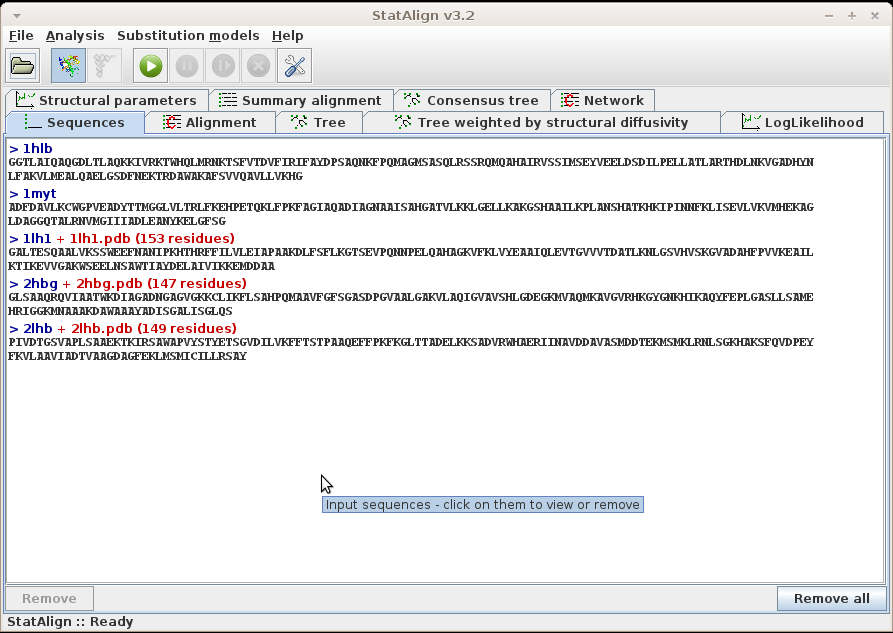
Sequences (and other data)
Sequences can be loaded from the menu, following
File → Add sequence(s)..., or by selecting the
link in the welcome page.
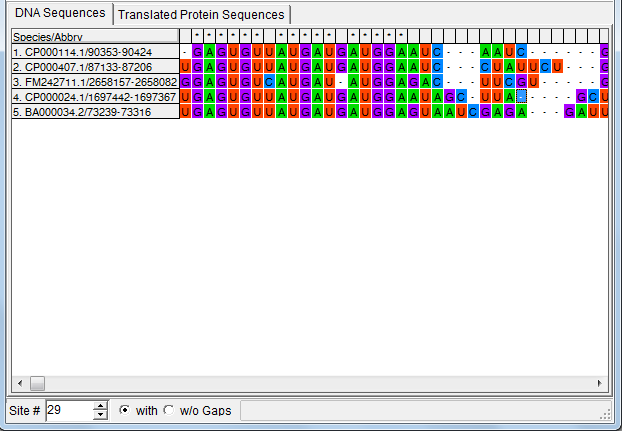
The loaded sequences are shown on the Sequences tabulated panel,
in standard Fasta format. For easier visualisation, the name of the
sequences are highlighted with dark blue, and printed with different
type of fonts.
To remove a sequence from the list, click into the desired sequence and
then to the Remove button. Adding sequences can be done simply
by following File → Add sequence(s)....
The sequences are automatically recognised if they are nucleotide or
protein sequences, and a default substitution model is associated to them.
If the sequences are not recognised, an error message is shown, indicating
what are the unknown characters in the input sequences.
When other types of data are added and associated with each sequence, for example
protein structures, these will also appear
alongside the sequence name.
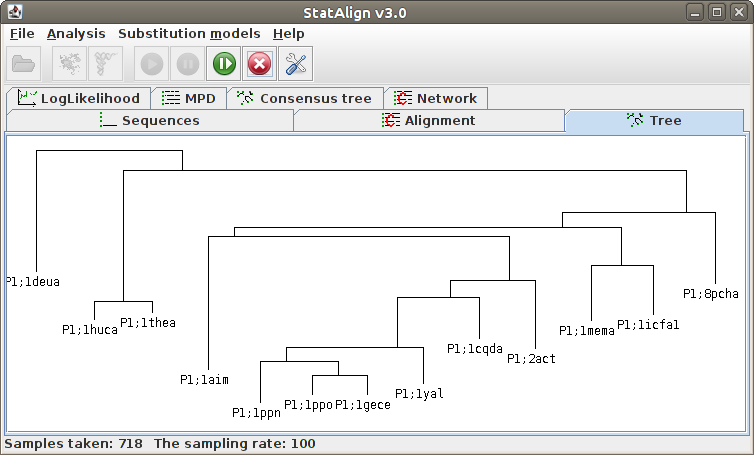
[
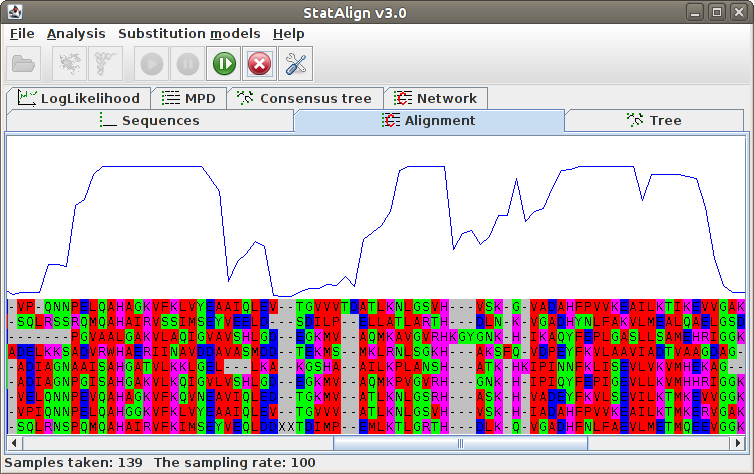
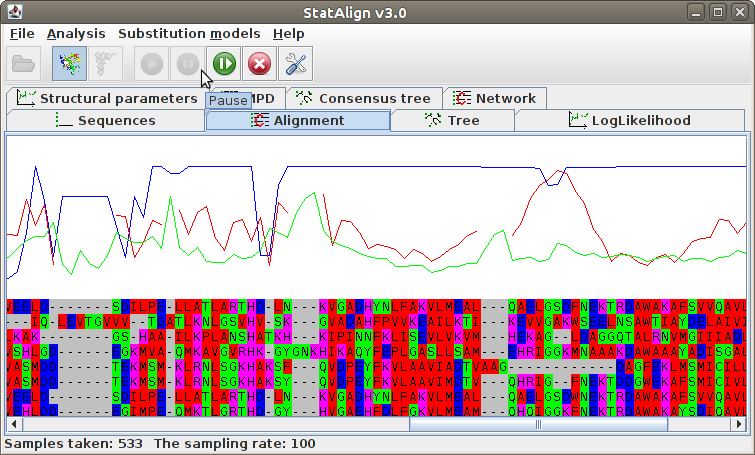
Maximum Posterior Decoding alignment
The program estimates summarises the alignment samples so far to produce a
type of consensus alignment, using maximum posterior decoding (MPD)
 .
The posterior probabilities for each column are printed on top of the alignment, indicating
the reliability of the corresponding MPD alignment column.
.
The posterior probabilities for each column are printed on top of the alignment, indicating
the reliability of the corresponding MPD alignment column.
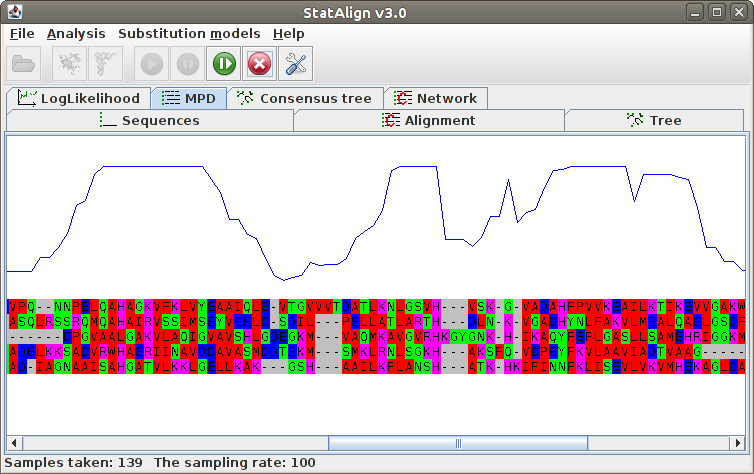
The MPD alignment is estimated using only alignment samples from the after-burn-in
period, hence the alignment is not shown during the burn-in phase in this panel.
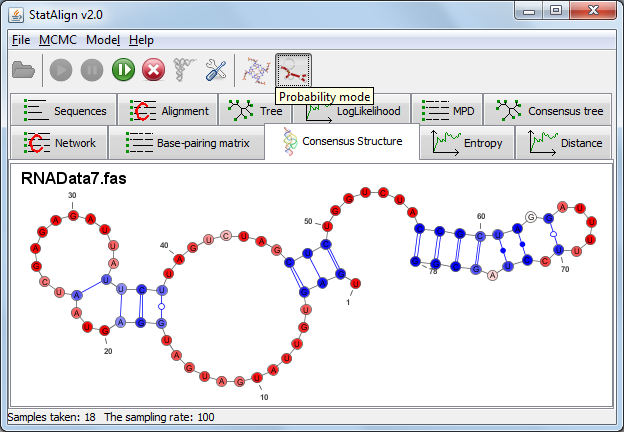
[
Introduction
The RNA plugin performs secondary structure predictions from multiple alignment samples generated by StatAlign. Both a SCFG approach (PPfold)
and a thermodynamic (RNAalifold) are available.
The advantage of this method of secondary structure prediction is that it considers multipled sampled alignments instead of just a single fixed alignment, as is typically the
case with comparative secondary structure prediction.
Here we describe the options available through the graphical interface.
Options available through the command line interface can be listed by running
java -jar StatAlign.jar -help:rnaalifold
java -jar StatAlign.jar -help:ppfold
The following flowchart summarises what StatAlign does when RNA mode is enabled:
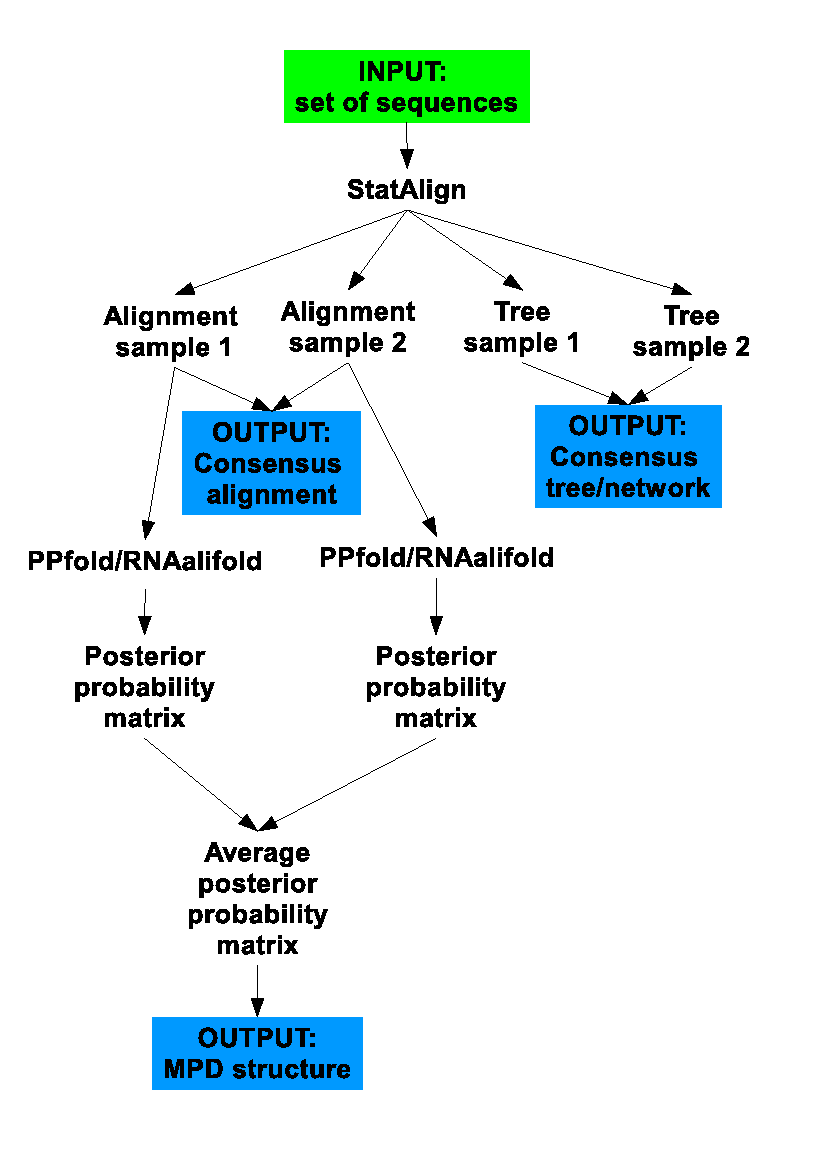 [
[
RNA mode settings
Activating the RNA mode brings up a settings dialog. Two different prediction methods are available for predicting structures from multiple alignment samples.
The first is an SCFG (Stochastic Context-Free Grammar) approach as implemented in PPfold ("Sampling and averaging (PPfold)").
Two predictions are produced using this approach: the standard sampling and averaging prediction, which produces a secondary structure from an averaged base-pairing probability matrix and a consensus evolutionary approach, which provides an information entropy value that takes into account contributions from alignment samples in addition to providing a secondary structure prediction.
A thermodynamic method ("Sampling and averaging (RNAalifold)")
can also be used, this method requires that user specify the RNAalifold executable (this shouldn't be necessary on Windows). This method allows the user to specify various
folding parameters such as the folding temperature and the genome conformation. The StatAlign GUI, however,
only provides some of these options. By running StatAlign from the command-line, additional RNAalifold parameters can be specified (see the RNAalifold manpage for a list of parameters).
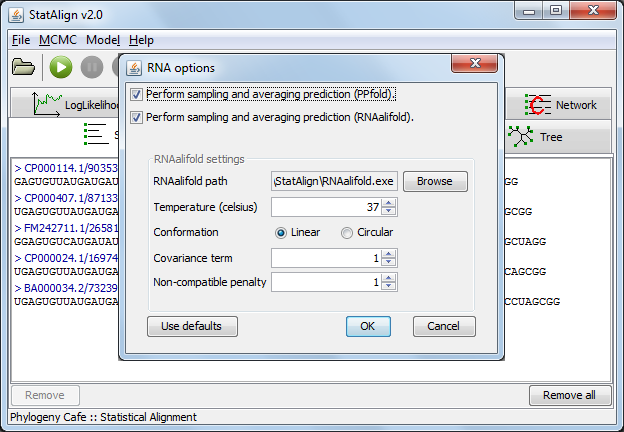
The GUI RNAalifold options are as follows:
- Temperature - Rescale energy parameters to this temperature.
- Conformation - Specify whether the sequence is a linear or circular, most often the sequence is linear.
- Covariance term - Set the weight of the covariance term in the energy function.
- Non-compatibility penalty - Set the penalty for non-compatible sequences in the covariance term of the energy function.
[
Once the RNA mode is activated it will run alongside the normal StatAlign run.
[
Information entropy
Information entropy is a measure of the spread of a probability distribution. The PPfold method is a SCFG method (Stochastic Context-Free Grammar method)
used to predict secondary structures. This SCFG method places a probability distribution on secondary structures which means that the information entropy of this
probability distribution can be calculated (Anderson et. al. (2012), in preparation, available upon request).
A low entropy corresponds to a probability distribution where the probability mass is concentrated on
a few structures, whereas a high entropy indicates that the probability mass is spread over many structures - this is undesirable as it can be an
indication that there are many alternative secondary structures which are almost as equiprobable as the most probable structure.
The information entropy calculation provided in StatAlign has been extended to reflect that fact that the PPfold sampling and averaging method samples over
alignments, instead of using a single fixed alignment.
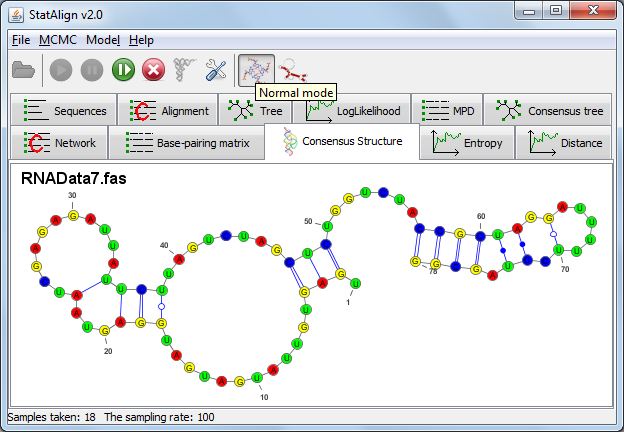
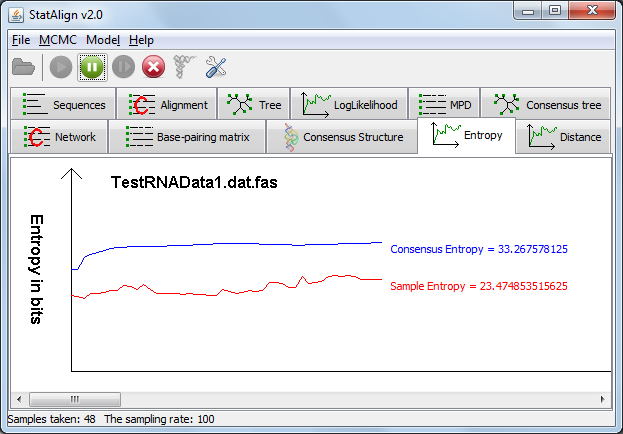
In the 'Entropy' tab, two line graphs are displayed depicting the 'Sample Entropy', which is the information entropy of the PPfold structure predictions
on individual alignment samples measured in bits, and 'Consensus Entropy' which is the extended information entropy that takes into account alignment space.
The 'Consensus Entropy' should approach a constant as the number of samples increases.
The final information entropy value can be found in the ".info" file on the header line corresponding to the consensus evolutionary prediction.
 [
[